Objective¶
The mission of the project is to provide food aid to people stuck under blue tarps in Haiti due to the earthquake. The data contains pixel images captured from the sites. The pixel correspond to various colors and our goal is to identify the blue tarps. The shape of the data for both the datasets is given below.
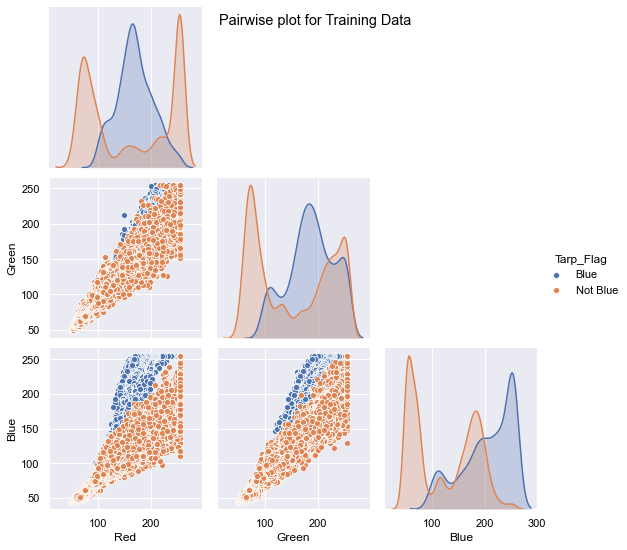
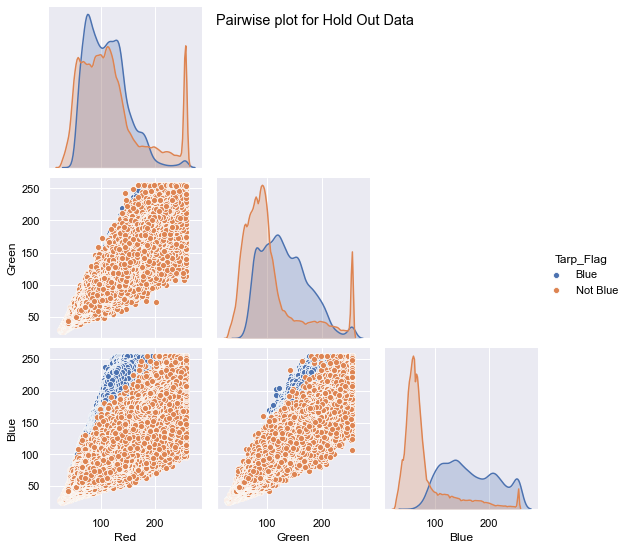
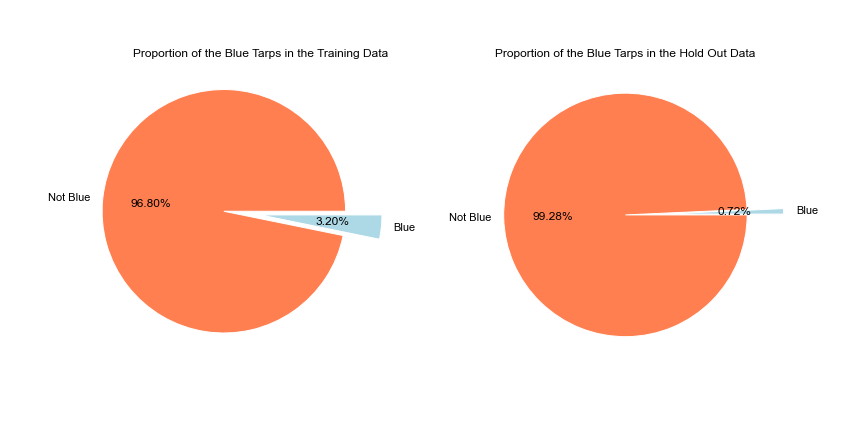
The training data has around ~63k records which contain 3.2% of Blue Tarps and the Hold Out dataset contains ~2million records and has less than 1% blue tarps.
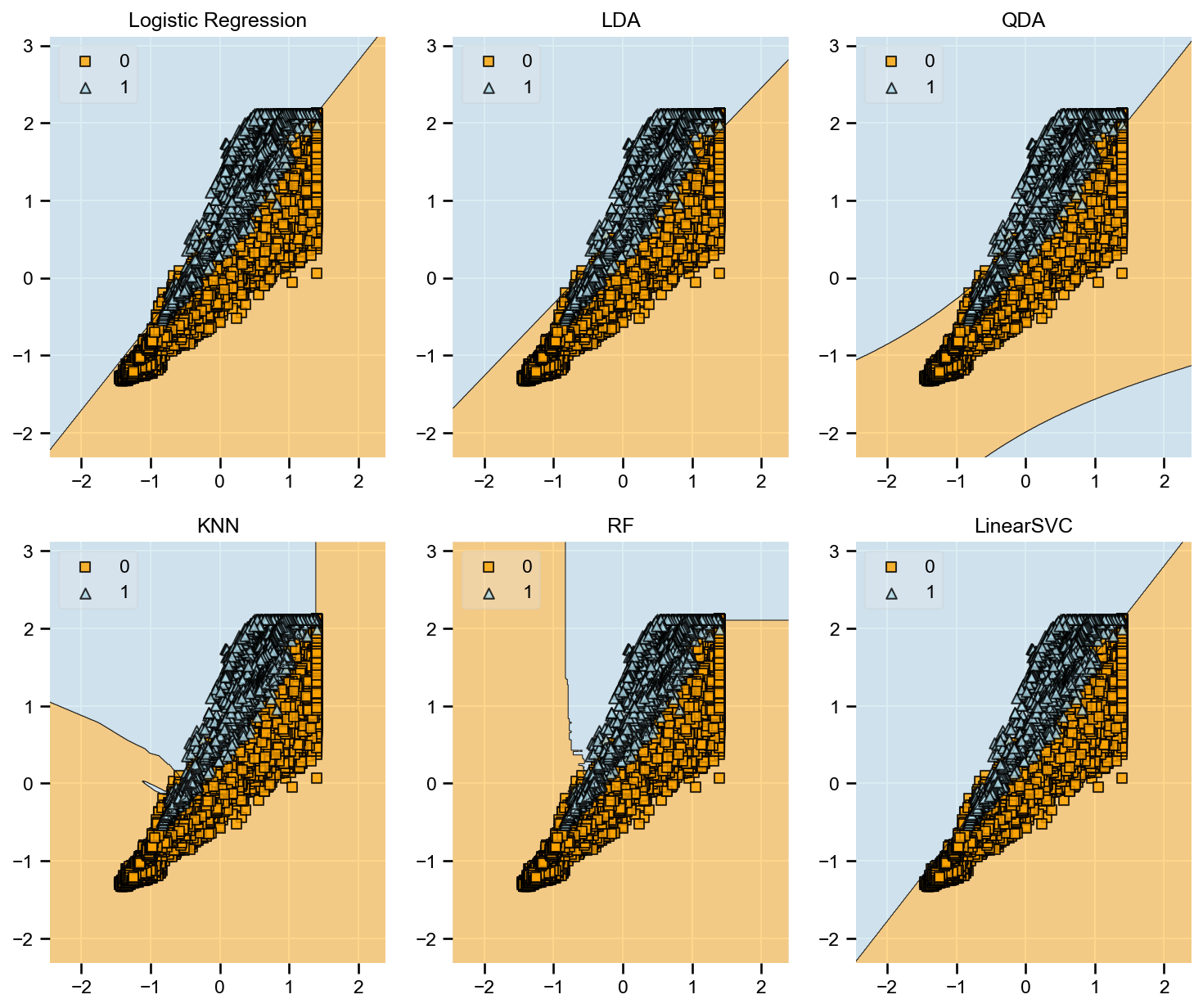
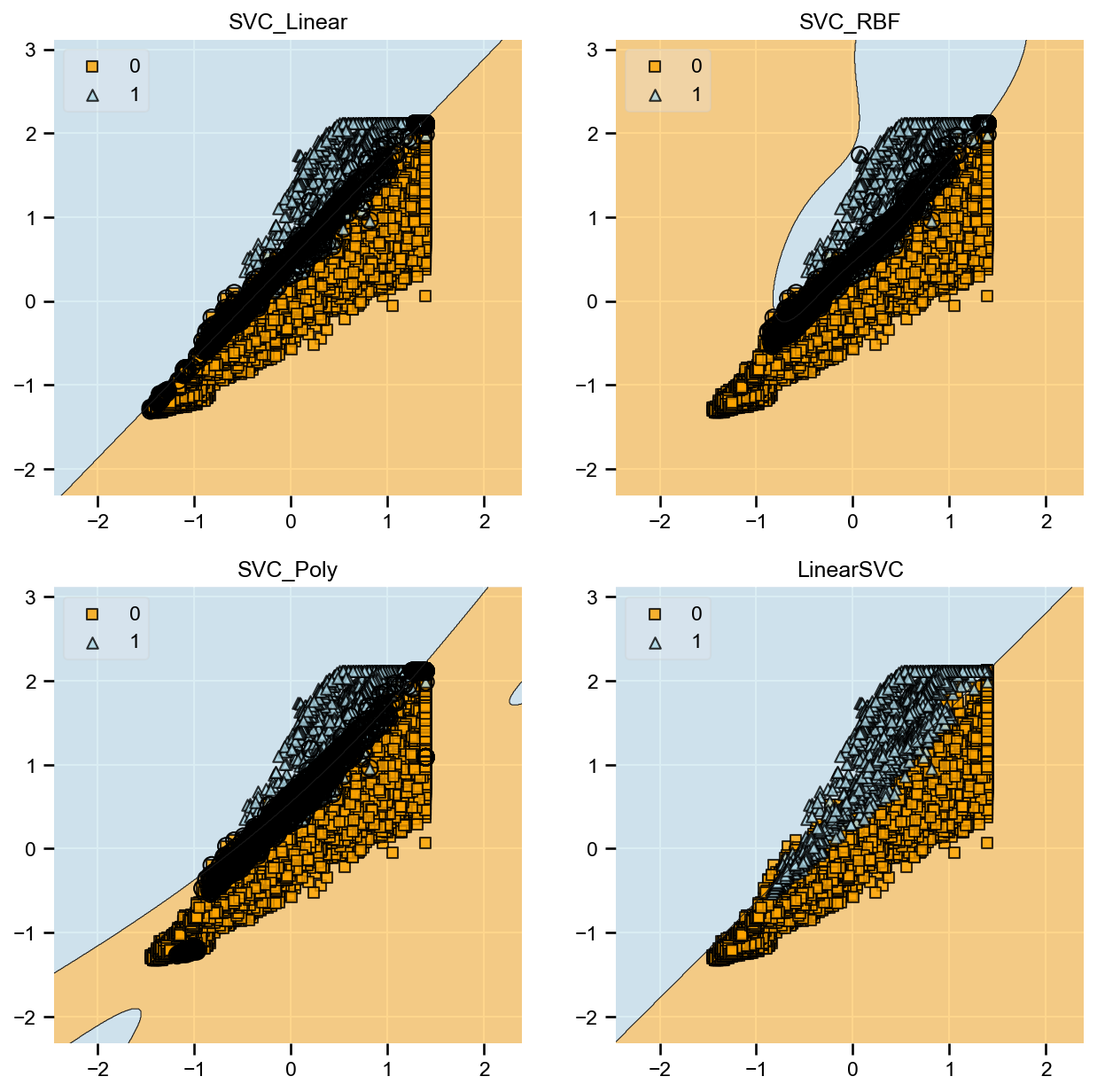
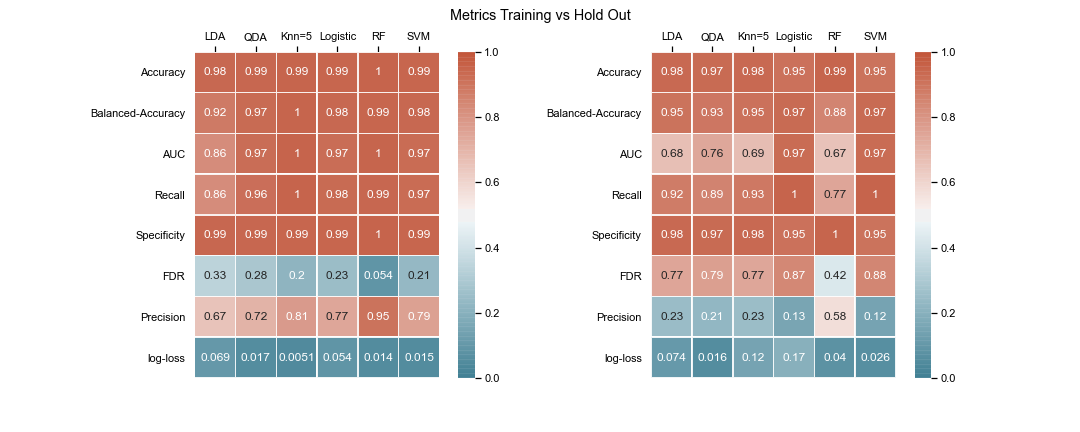
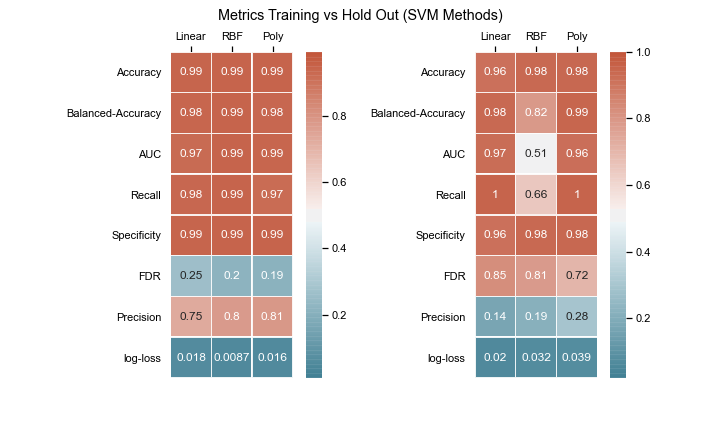
The goal is to compare various machine learning algorithms and discuss the metrics to identify the best model. The list of models being compared are:
- Linear Discriminant Analysis (LDA)
- Quadratic Discriminant Analysis (QDA)
- K Neighbor Classifier (KNN)
- Logistic Regression (LR)
- Random Forest (RF)
- Linear SVC
- SVM (Linear Kernel)
- SVM (Radial Kernel)
- SVM (Poly Kernel)
Approach:
Since our main mission is to save lives, we need to identify the Blue Tarps to save the people. However, such missions are limited by resources, time and budget and in many cases, we cannot acheive all the three. It is very crucial to predict the blue tarps accurately as much as predicting the non-blue tarps because we do not want to waste our time and resources on False Positives. Similarly, we also do not want to miss saving lives because of False Negatives.
Some important definitions for terms used throughout the document for quick reference:
True Positives (TP): These are cases in which we predicted YES (Blue Tarps), and they are Blue Tarps.
False Positive (FP): predicted YES, but not Blue Tarps. ("Type I error.")
True Negatives (TN): predicted NO (Not Blue Tarp) and are not
False Negative (FN): predicted NO, but Blue Tarps ("Type II error.")
Accuracy: Overall, how often is the classifier correct
$TP+TN/(TP+FP+TN+FN)$
Balanced Accuracy: Normalizes true positive and true negative predictions by the number of positive and negative samples, respectively, and divides their sum by two
$Balanced Accuracy = ((True Positive Rate + True Negative Rate)/2)$
AUC: Area under the curve. For this document it is the area under the precision-recall curve
Precision: Precision summarizes the fraction of examples assigned the positive class that belong to the positive class
$Precision = TP / (TP + FP)$
Sensitivity or Recall: Recall summarizes how well the positive class was predicted and is the same calculation as sensitivity
$Recall = $TP / (TP + FN)$
F Score: This is a weighted average of the true positive rate (recall) and precision. Special cases of F score based on beta which defines the weight of precision and recall are given below:
- F0.5 - Measure ($beta=0.5$): More weight on precision, less weight on recall
- F1 - Measure ($beta=1.0$): Balance the weight on precision and recall
- F2 - Measure ($beta=2.0$): Less weight on precision, more weight on recall
False Discovery Rate: The expected proportion of type I errors. A type I error is where you incorrectly reject the null hypothesis; In other words, you get a false positive.
Logarithmic loss (log-loss): Log-loss measures the performance of a classification model where the prediction is a probability value between 0 and 1. Log loss increases as the predicted probability diverge from the actual label. Log loss is a widely used metric for Kaggle competitions. Lower the log-loss value, better are the predictions of the model.
The below measures have been taken to balance the class imbalance, False Positives, False Negatives and to accurately predict True Positives and True Negatives:
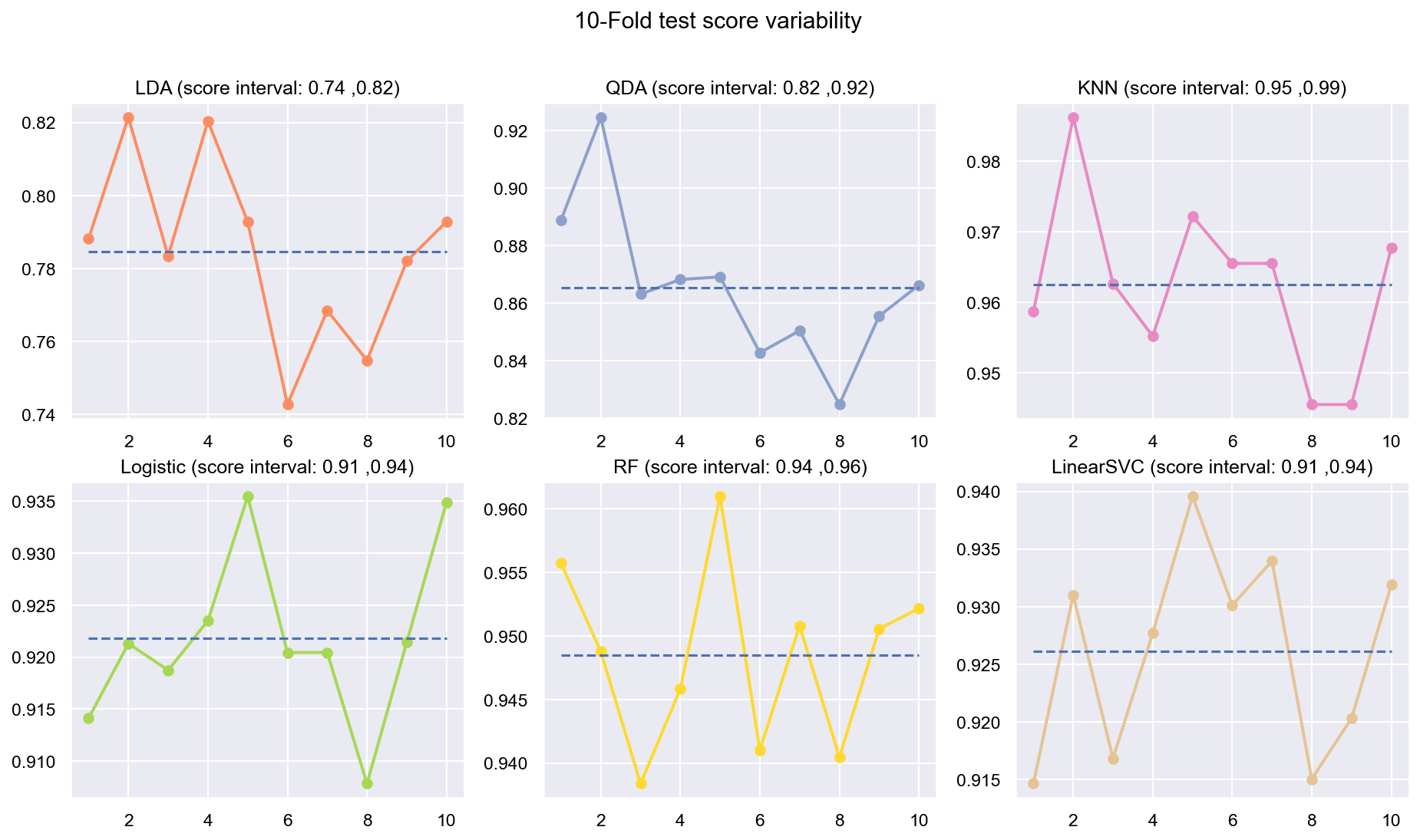
- Use Stratified K-FOLD to represent both the classes well in cross-validation
- Use class_weight= ‘balanced’ or equivalent for methods wherever possible to balance both the classes while fitting the data
- Use balanced accuracy, precision-recall curve AUC, precision, recall, f-score, log-loss as major metrics to determine the performance of an algorithm.
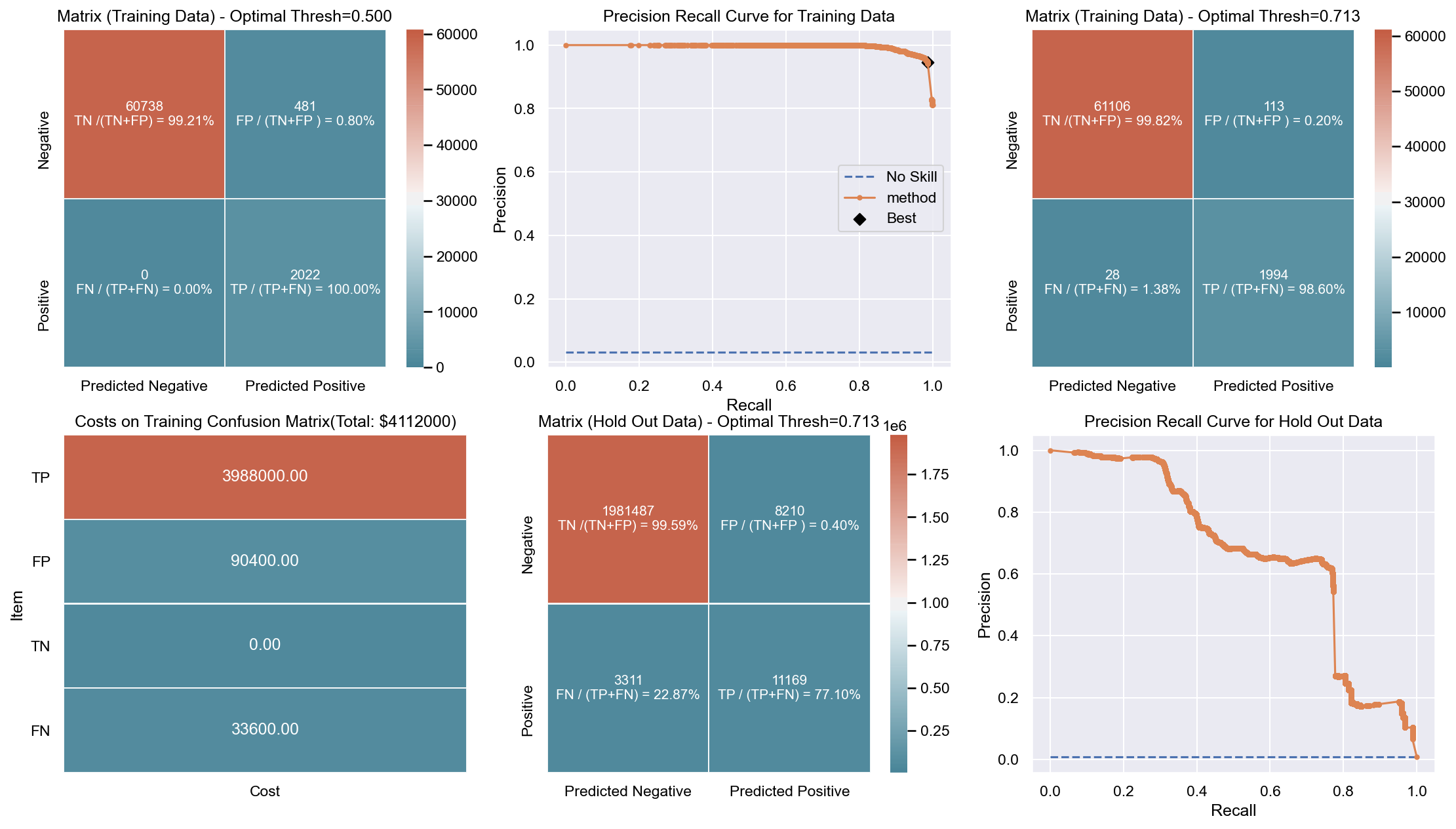
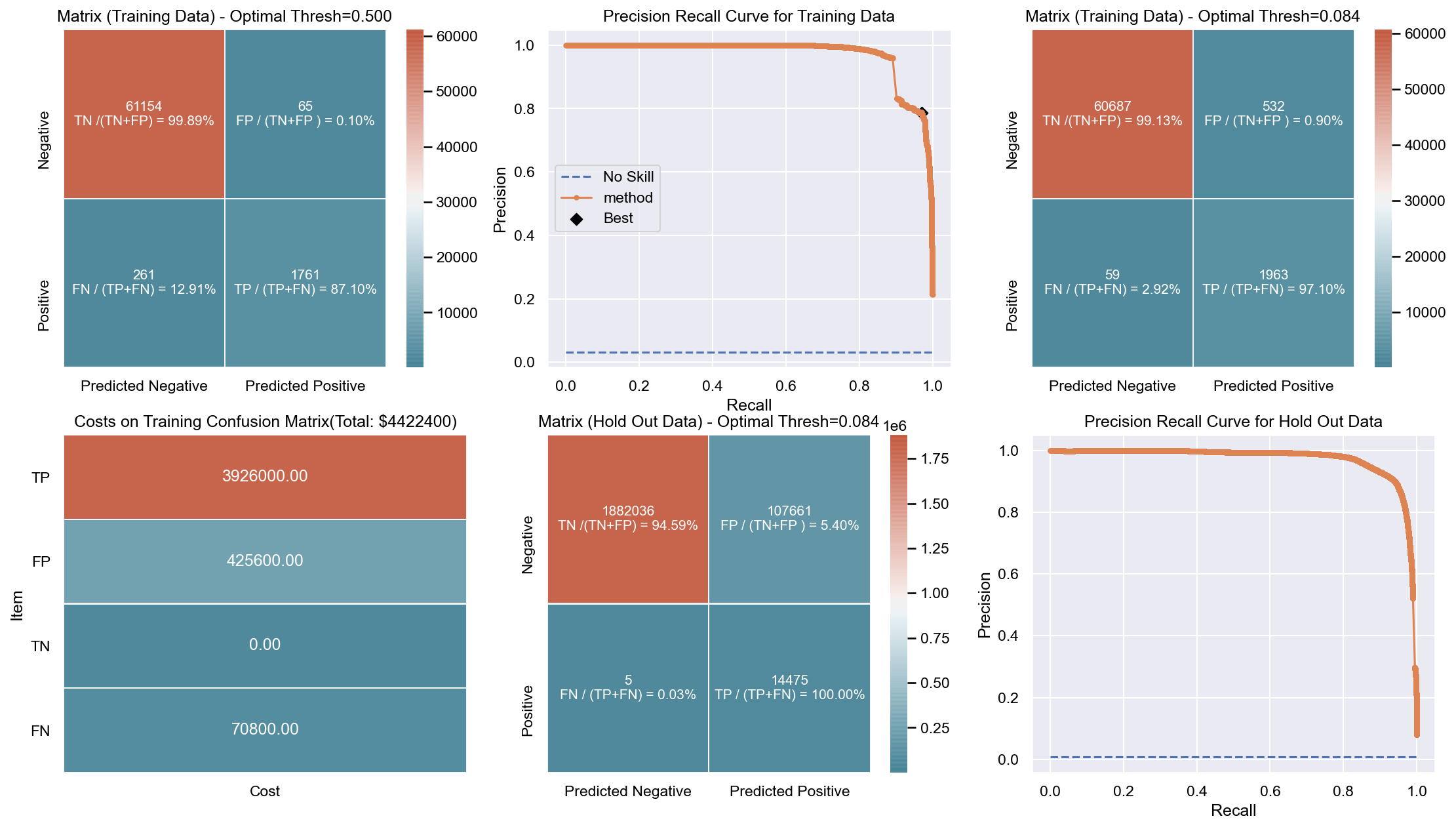
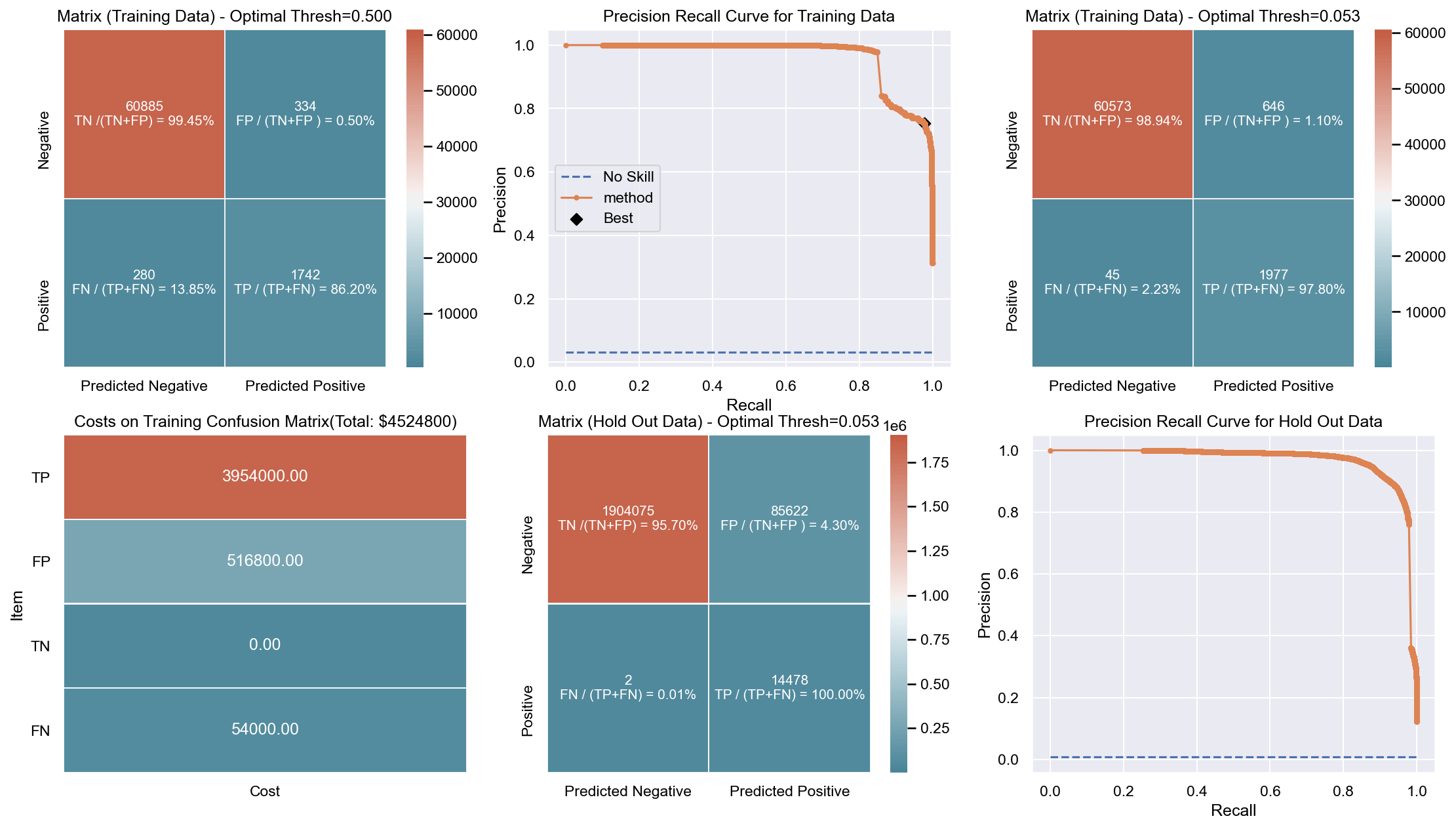
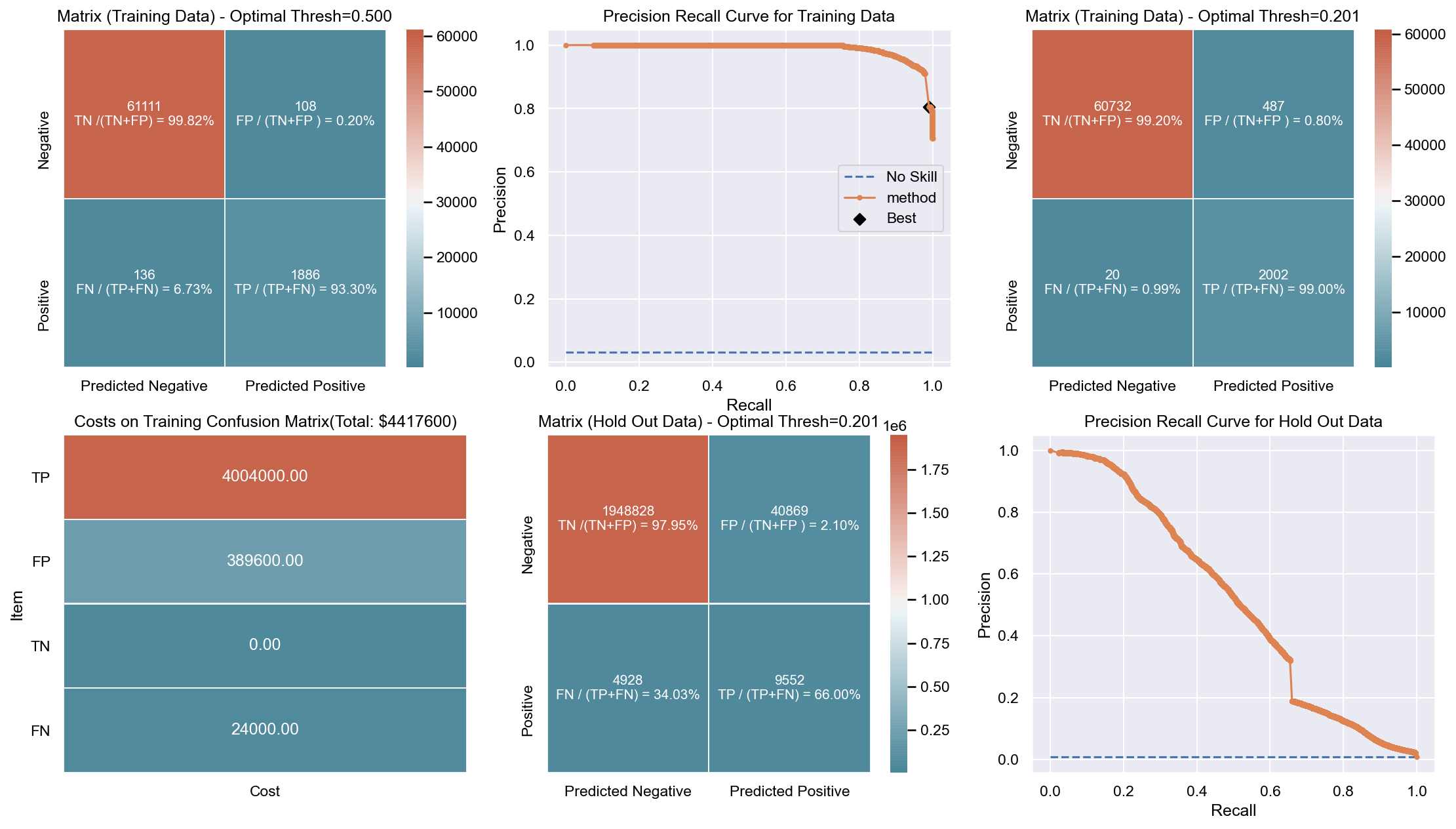
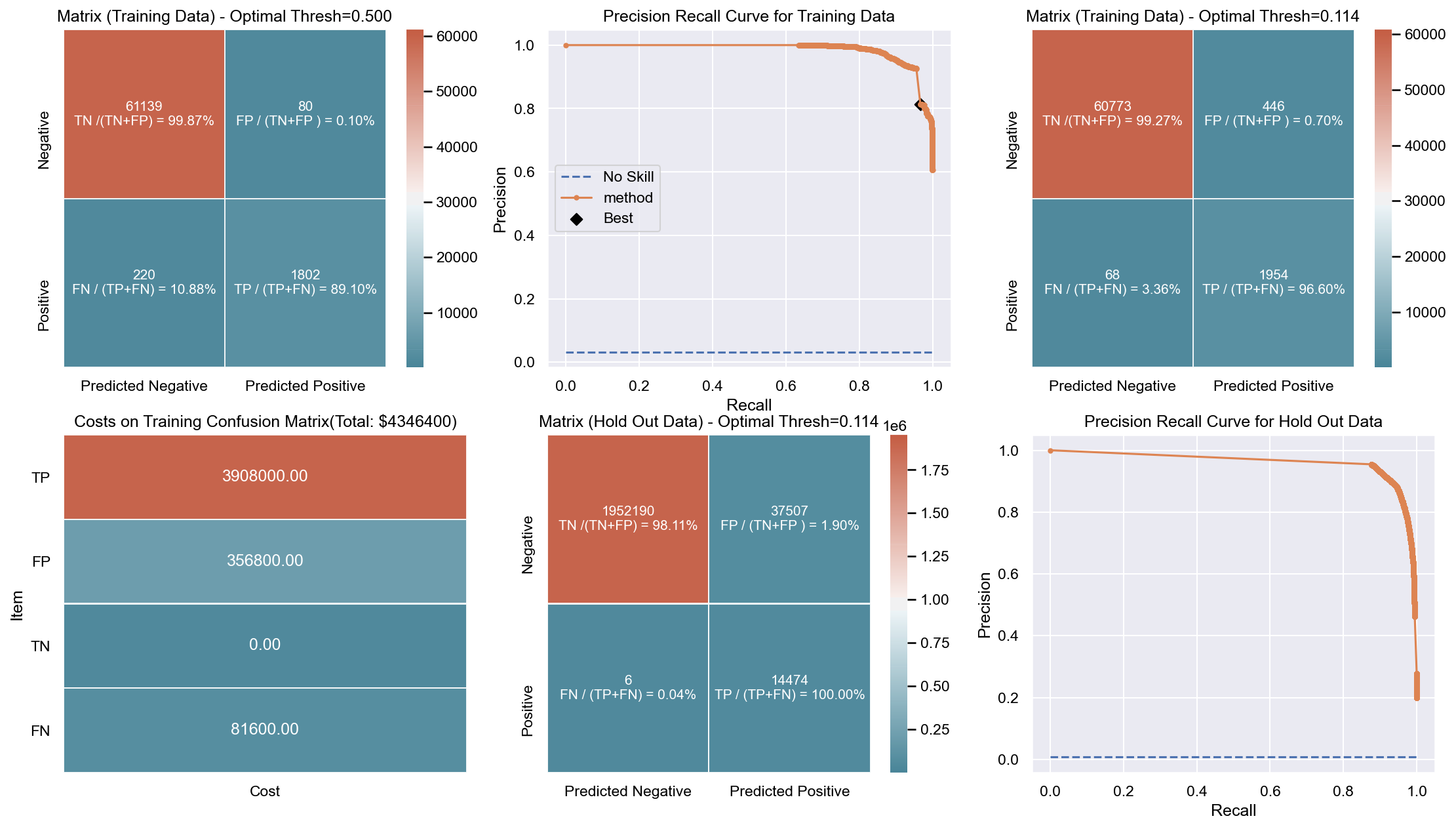
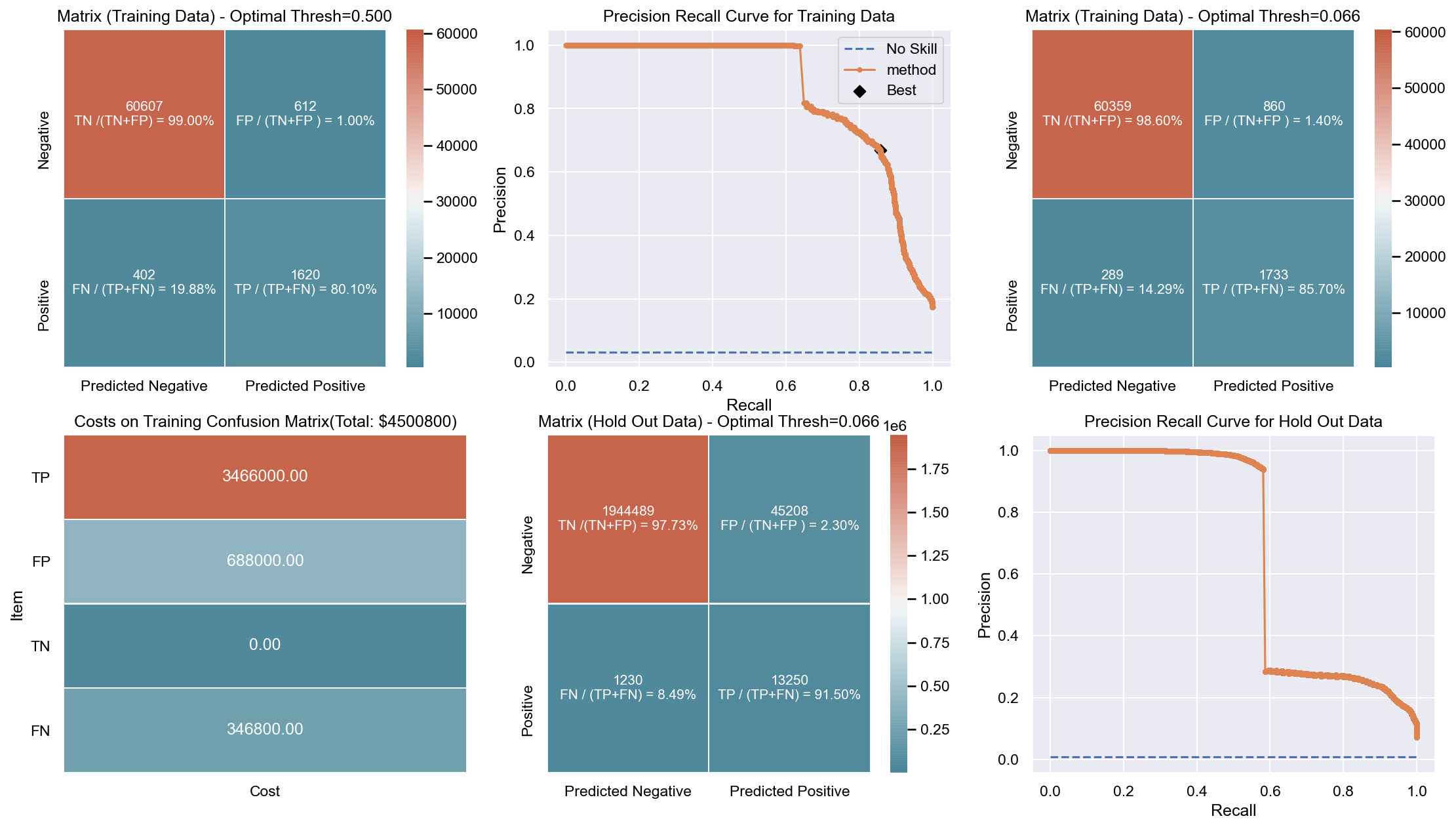
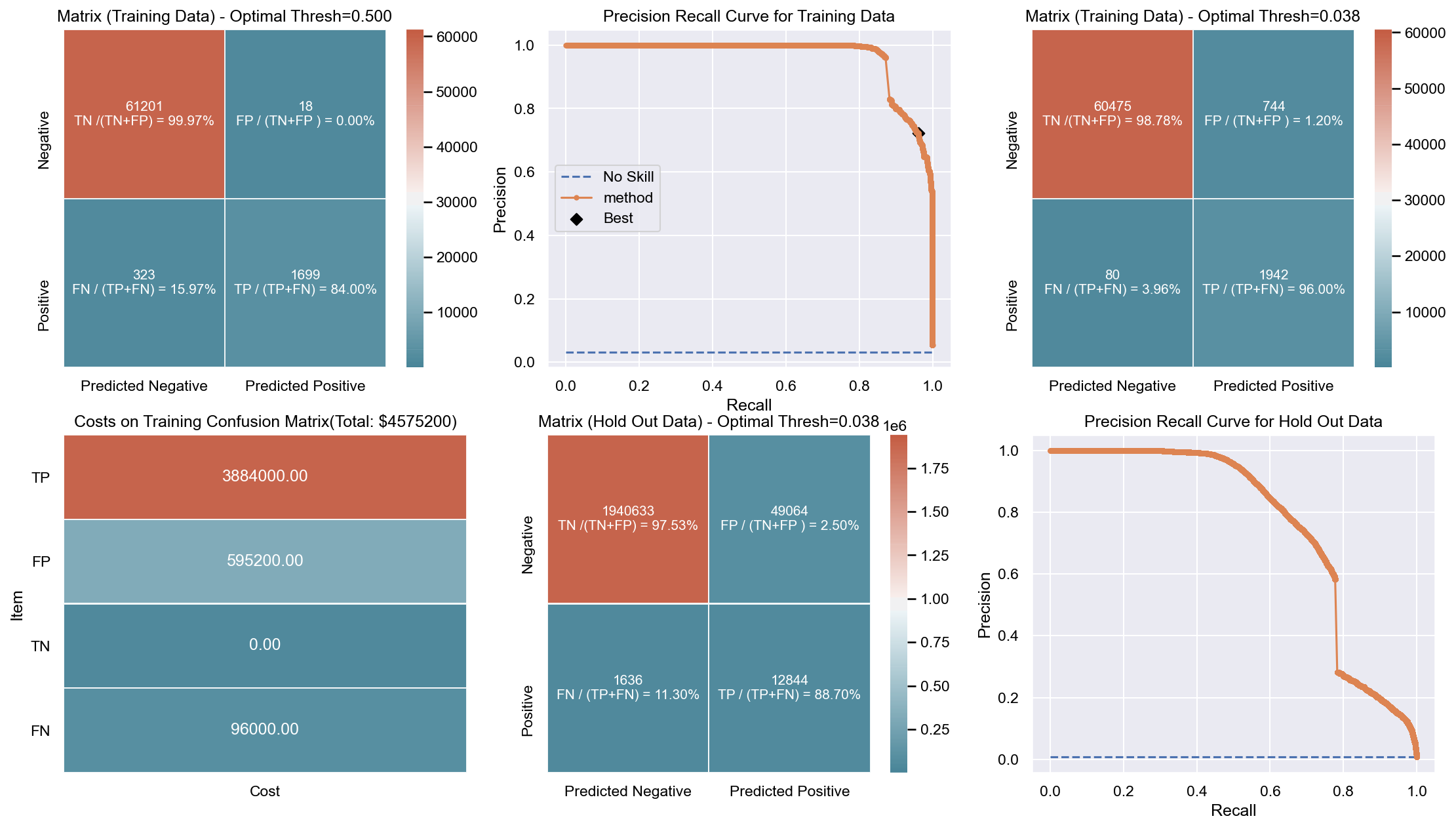
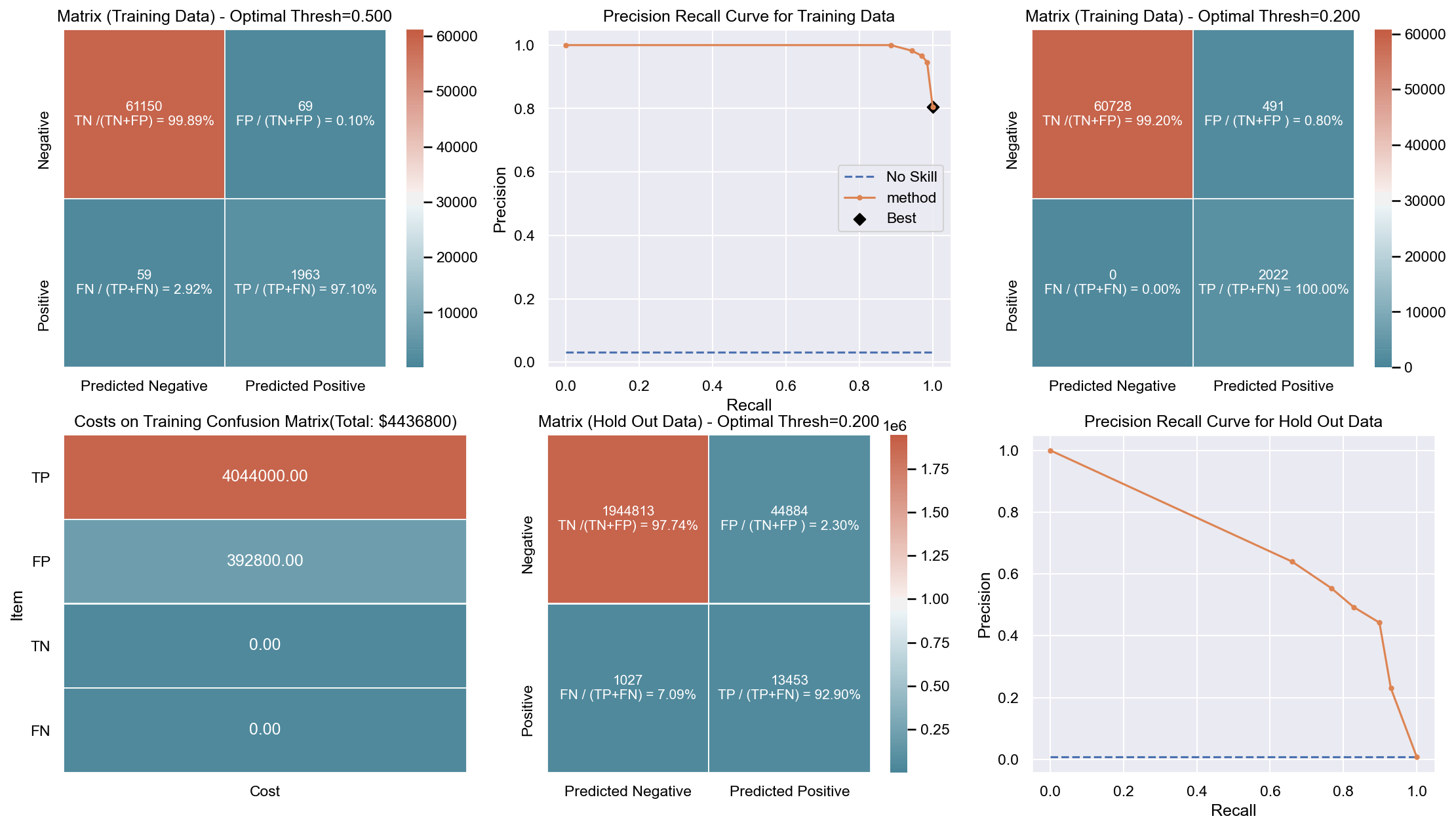
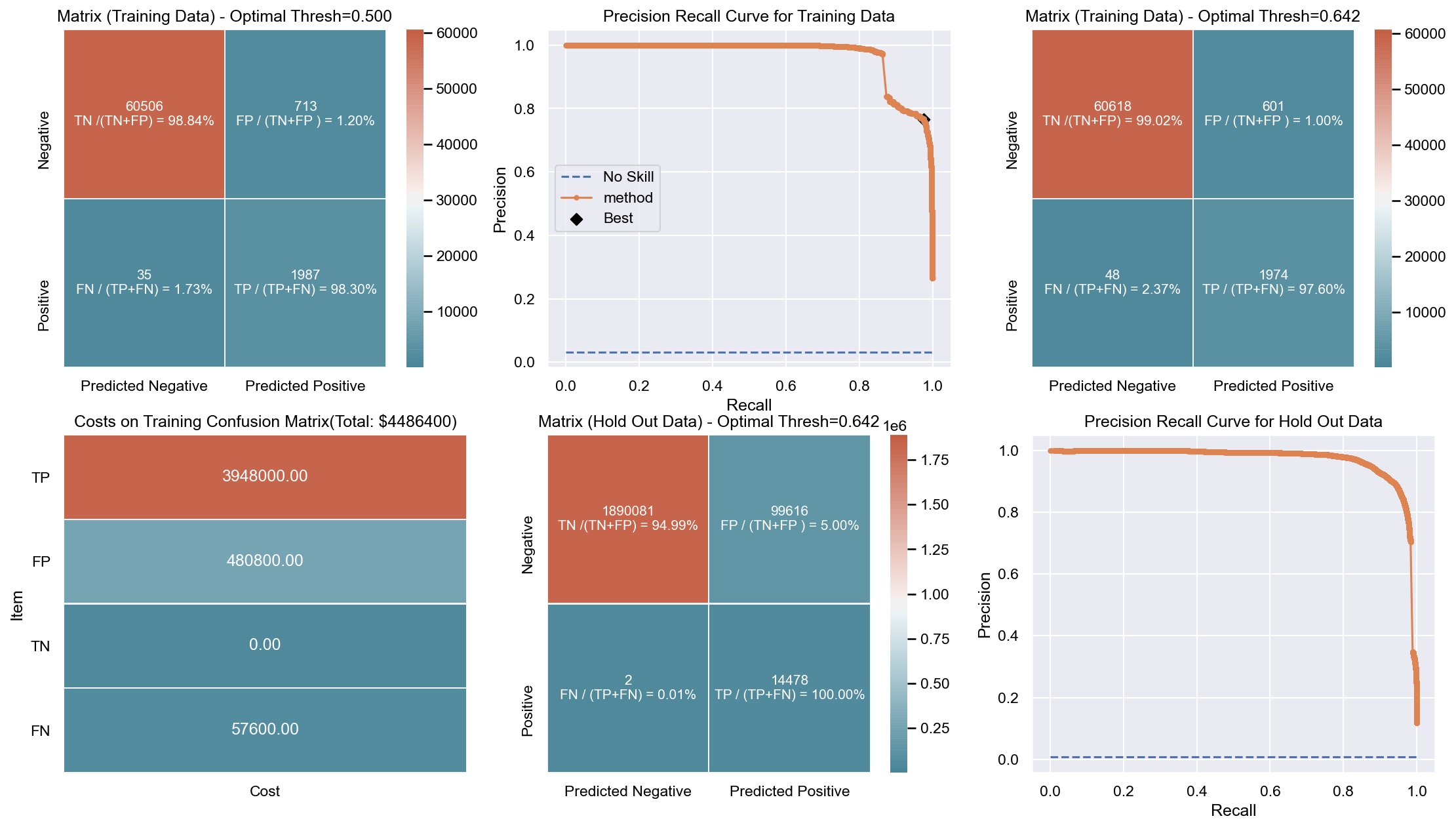
Due to the huge imbalance in data some of the metrics can be mis-leading and especially because we are interested in predicting the positive class which is a minority. The method used to score the cross-validation is ‘fbeta’. Precision and recall matter because in this scenario we also care about resources, time and budget along with prediction capability. Therefore, giving he correct weight to balance precision and recall is critical. To demonstrate the capability of algorithms I decided to use beta as 2 as I personally care more about FN. I created an interactive Jupyter dashboard which can be used to plug in cost values for TP, FP, TN, FN and provide a beta value to see the change and shift in results of costs for various algorithms which can be found here. This dashboard will help the parties make an informed decision by providing the metrics and costs. This can be scaled to pass the algorithm or other parameters which might affect the decisions.
Though it has been challenging to determine the weight for precision and recall, it is hard to ignore the fact that costs will be driving decisions in such a scenario. Therefore, for demonstration purpose I have considered the below hypothetical costs (TP,FP,TN,FN - per person):
Budget: $\$4million$ <br> __TP:__ $\$2000$
FP: $\$800$ <br> __TN:__ $\$0$
FN: $\$1200$
The rational for the costs above are driven by a general cost analysis model. When there is a budget, we assign a cost to everything. Therefore, though the main mission is to save lives it comes with a cost. There is at least cost of employees, fuel and food. Therefore, identifying true positives is more expensive than other categories. Because our mission is still to identify as many Blue Tarps as possible, we have more cost on FN and somewhat less on FP. If we really predict a TN then we do not have any costs as there is no action. To simplify the analysis the goal to stay within $\$4million$, identify as many Blue Tarps as possible. The cost analysis done is just an example to demonstrate an idea of how it can be approached.
 |
| 
 '
'